Abstract
Chromosomal translocations and inversions that create fusion genes are important causal events in AML. 7 recurrent fusion genes define AML entities in the 2016 revision of the WHO classification of hematologic malignancies. We have developed a comprehensive diagnostic AML platform based on next-generation mRNA sequencing (RNAseq). The adaptable integrated bioinformatic pipeline HAMLET (Human AML Expedited Transcriptomics) analyses the raw data simultaneously for fusion transcripts and for sequence variants and expression levels of genes with relevance to AML pathogenesis, prognosis, or actionability.
Poly(A)+ RNA from 100 cryopreserved AML samples with a blast count of 10-99% (median: 75%) from 96 patients (3 diagnosis-relapse pairs, 1 de novo AML and subsequent tAML) was sequenced at a depth of 59x106 paired-end reads/sample with median read length of 126 bp in an ISO17025-accredited Illumina HiSeq 2500 pipeline. Fusion transcripts were called by STAR-Fusion and FusionCatcher. HAMLET detected the recurrent fusion transcripts RUNX1-RUNX1T1, CBFB-MYH11, PML-RARA, ERG-FUS, DEK-NUP214, and KMT2A-MLLT3 plus the variants KMT2A-MLLT4, KMT2A-MLLT1, and KMT2A-MLLT6 with 100% sensitivity and 100% specificity compared to metaphase cytogenetics, FISH, and RT-PCR (Griffioen, ASH 2016).
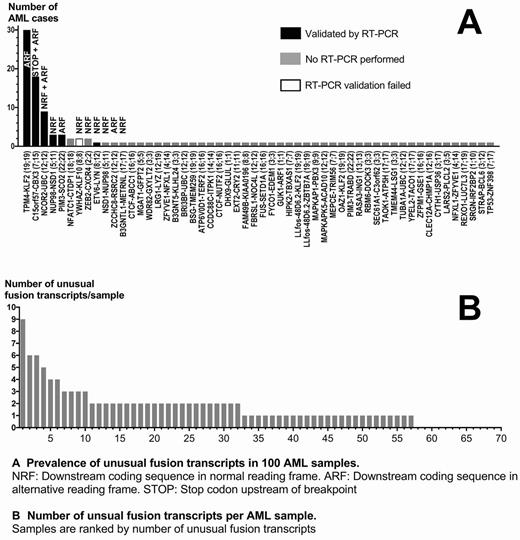
In addition, HAMLET identified fusion transcripts from 54 combinations of genes in 57 AML samples that lacked corresponding chromosomal aberrations by metaphase cytogenetics. 14 "unusual" fusions involved genes located on different chromosomes; 40 fusions represented intrachromosomal events. 8 fusions were detected recurrently, i.e. in at least 2 cases (Fig. A). 32 AML carried at least 2 unusual fusions, with a maximum of 9 different fusions in an AML with a complex karyotype (Fig. B). Unusual fusions were not correlated with classical recurrent AML fusion genes. The 5 most prevalent fusions and an ETV6-LYN fusion were successfully validated in a total of 18 AML by RT-PCR. The only fusion that could not be validated was a YWHAZ-KLF10 fusion (2 cases).
The most prevalent RT-PCR-validated fusion (seen in 30 AML cases) has been described in ALL (Roberts, 2012; Safavi, 2015) and was an intrachromosomal TPM4-KLF2 fusion between the tropomyosin 4 and an alternative reading frame (ARF) of the transcription factor Kruppel-like factor 2 genes. Unexpectedly, we detected this fusion in all 3 tested normal bone marrows (BM) but not in EBV-transformed B cells. The hitherto unvalidated C15orf57-CBX3 fusion may be generated by retrotransposition (Schrider, 2013) and was present in 18 AML as well as in 2/3 normal BM and 2/3 EBV B-cells with CBX3 in ARF. The intrachromosomal NCOR2-UBC fusion (9 AML) is listed in the database ChimerDB without further characterization and involves the genes of Nuclear corepressor 2 and Ubiquitin C in the normal reading frame (NRF). The NUP98-NSD1 fusion (3 AML) with NSD1 in NRF is frequently found in hematological malignancies including AML and originates from a cryptic t(5;11) due to telomeric positions of both genes. The PIM3-SCO2 fusion between the serine/threonine kinase PIM3 and an ARF of Cytochrome C oxidase assembly protein can originate from a 0.6 Mb inv(22) in chronic neutrophilic leukemia (Menezes, 2013) and occurs frequently in pediatric AML (Kuhn, 2013). The well-characterized ETV6-LYN fusion (1 AML) fusing the TEL1 oncogene to the SRC family kinase LYN in NRF originates from a complex translocation with inversion involving chromosomes 8 and 12 (Telford, 2016) and has oncogenic activity by activation of STAT5 (Takeda, 2011).
Fusion transcripts without recognizable chromosomal abnormalities on metaphase cytogenetics occur frequently in AML and involve actionable targets. The upstream sequence can be fused to the normal or alternative reading frame of the downstream coding sequences and involve potential or validated proto-oncogenes. Apart from cryptic translocations or inversions, trans-splicing is suggested as an alternative mechanism by the presence of some fusion genes in non-malignant hematopoietic cells. Trans-splicing without DNA rearrangement can occur physiologically and alter cellular development towards potential premalignant differentiation (Yuan, 2013). Increasing use of RNAseq technology in routine AML diagnostics, e.g. by HAMLET, will permit to unravel the prognostic significance of unexpected fusion transcripts and to guide novel targeted therapies.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal